 | 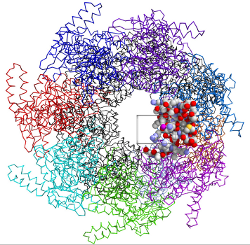 | 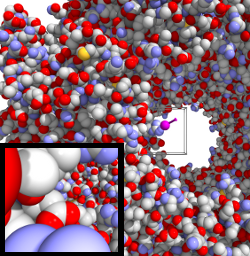 |
| HaptiMol ISAS enables users to explore the solvent accessible surface with a haptic feedback device. The user manipulates the magenta probe sphere and exploration is achieved via the Navigation Cube, shown by the cube. | The backbone of the molecule can be rendered to give the user a more familiar view of the molecular structure. Only atoms surrounding the Navigation Cube are rendered with ray-casting. | A third person view of the probe interacting with GroEL. A view from the probe sphere (Probe Camera) in the direction of the magenta viewfinder can be obtained. Inset bottom-left: A view from the probe camera. |
HaptiMOL
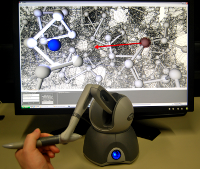
The HaptiMOL suite enables interaction with protein structures using force feedback, through the use of a haptic feedback device.
HaptiMOL DockIT
DockIT is a tool that has a unique set of physical and graphical features for interactive molecular docking. It enables the user to bring a ligand and a receptor into a docking pose by controlling the ligand position, either with a mouse and keyboard, a haptic device or VR touch controllers. The Oculus Rift, RiftS or Meta Quest 2 (with link cable) are supported).
Receptor flexibility can also be incorporated. In order to model receptor flexibility we use the method of linear response for which we determine the variance-covariance matrix of atomic fluctuations from the trajectory of an explicit-solvent Molecular Dynamics simulation of the ligand-free receptor molecule. Atomic interactions are modelled using molecular dynamics-based force-fields with the force on the ligand being felt on a haptic device. Real-time calculation and display of intermolecular hydrogen bonds and multipoint collision detection either using maximum force or maximum atomic overlap, mean that together with the ability to monitor selected intermolecular atomic distances, the user can find physically feasible docking poses that satisfy distance constraints derived from experimental methods. With these features and the ability to output and reload docked structures it can be used to accurately build up large multi-component molecular systems in preparation for molecular dynamics simulation.
A movie of DockIT is available.
The following images are screenshots from the software.
An OpenCL enabled GPU is required to run the software.
 |
 |
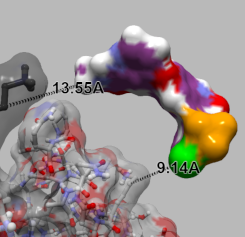 |
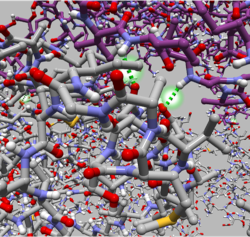
| GroEL and GroES. GroEL is rendered with ball-and-stick and semi-transparent molecular surface. GroES is rendered with the solid molecular surface. | The interatomic hydrogen bonds are depicted with green dashed lines. | Sorafenib and Braf. Dashed lines between atoms on each of the structures are shown to illustrate the distance between them. |
HaptiMOL ISAS
HaptiMOL ISAS enables the user to explore the solvent accessible surface of molecules using a probe sphere. A movie illustrating the features of the software can be downloaded by clicking movie. Version 3.0 of ISAS now includes improved visualisation using ray-casting and a Probe camera view. These features are shown in this video: Version 3.0.
The following images also illustrate some of the features of the software.
HaptiMOL ENM
HaptiMOL ENM enables users to apply forces to atoms in an elastic network model and to observe the resulting deformation. A movie illustrating the features of the software can be downloaded by clicking movie.
The following images also illustrate some of the features of the software.
 |  |  |
| The Navigation Cube is used to explore a protein. The dimensions of the cube are based on the usable workspace of the device (In this case the Novint Falcon). | Interacting with GroEL. The blue sphere represents the haptic probe, the Navigation Cube is displayed for aiding navigation. The haptic probe can be seen to illuminate the parts of the structure which are in close proximity to it using the Haptic Light. | The user has selected an atom and is applying a force to it be pulling the haptic device away from the selected atom. |
Protein Trajectory Viewer
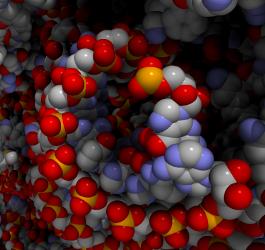
Protein Trajectory Viewer enables users to interactively explore high quality visualisations of trajectories of proteins in real-time. Ambient occlusion and shadows are calculated as the structures update to enhance the depth perception. Click here to download a movie illustrating Protein Trajectory Viewer.
You can load in PDB files containing multiple models or may load in a PDB file and AMBER NETCDF trajectory file separately.
The following images are screen shots taken from the software.
A CUDA enabled GPU is required to run the software.
 |  |  |
HaptiMOL RD
It is designed for Rigid Molecular Docking. A movie illustrating the software can be downloaded by clicking movie.
The following images also illustrate some of the features of the software.
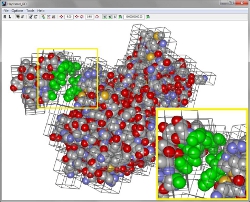 |
| Main image: Two proteins interact within Haptimol RD. Green colours denote the atoms that interact within a given cut-off distance. Inset: Zoomed in view of the interacting atoms. |
HaptiMOL FlexiDock (The features of flexidock are now included in DockIT)
The user “holds” a rigid ligand via a haptic device through which they feel interaction forces with a flexible receptor biomolecule. In order to model receptor flexibility we use the method of linear response for which we determine the variance-covariance matrix of atomic fluctuations from the trajectory of an explicit-solvent Molecular Dynamics simulation of the ligand-free receptor molecule.
A movie of HaptiMOL FlexiDock is available.